Note
Click here to download the full example code
1.6: 2D Visualization.¶
# Importing GemPy
import gempy as gp
# Importing auxiliary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
np.random.seed(1515)
pd.set_option('precision', 2)
Model interpolation¶
Data Preparation
data_path = 'https://raw.githubusercontent.com/cgre-aachen/gempy_data/master/'
geo_data = gp.create_data('viz_2d', [0, 1000, 0, 1000, 0, 1000], resolution=[10, 10, 10],
path_o=data_path + "/data/input_data/jan_models/model5_orientations.csv",
path_i=data_path + "/data/input_data/jan_models/model5_surface_points.csv")
Out:
Active grids: ['regular']
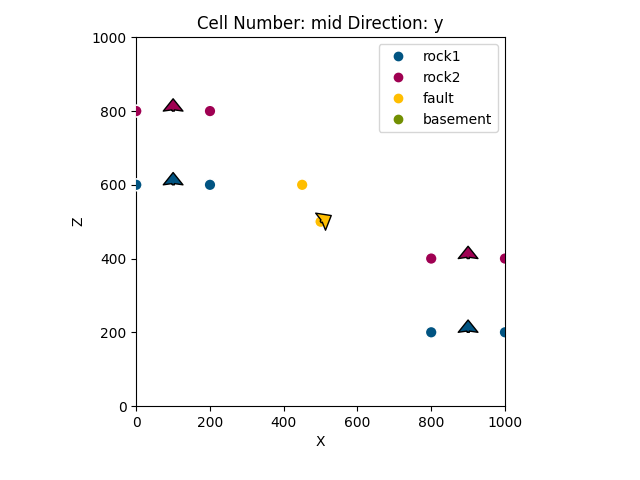
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcb8bab22e0>
geo_data.set_topography(d_z=(500, 1000))
Out:
Active grids: ['regular' 'topography']
Grid Object. Values:
array([[ 50. , 50. , 50. ],
[ 50. , 50. , 150. ],
[ 50. , 50. , 250. ],
...,
[1000. , 777.77777778, 601.70645773],
[1000. , 888.88888889, 547.79987061],
[1000. , 1000. , 531.67197073]])
section_dict = {'section1': ([0, 0], [1000, 1000], [100, 80]),
'section2': ([800, 0], [800, 1000], [150, 100]),
'section3': ([50, 200], [100, 500], [200, 150])}
geo_data.set_section_grid(section_dict)
gp.plot.plot_section_traces(geo_data)
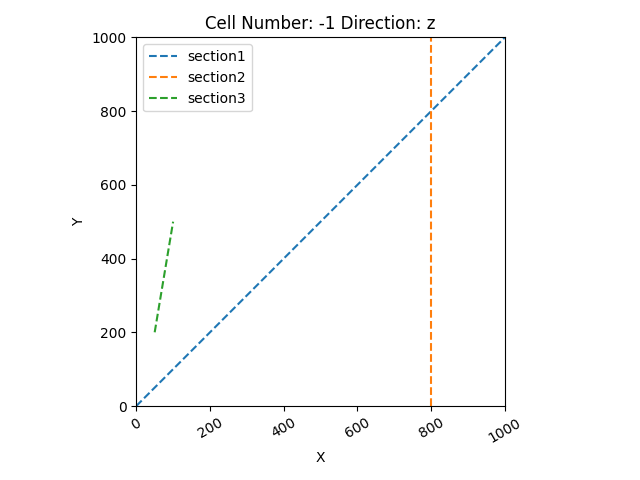
Out:
Active grids: ['regular' 'topography' 'sections']
<gempy.plot.visualization_2d.Plot2D object at 0x7fcb8912c580>
gp.set_interpolator(geo_data, theano_optimizer='fast_compile')
Out:
Setting kriging parameters to their default values.
Compiling theano function...
Level of Optimization: fast_compile
Device: cpu
Precision: float64
Number of faults: 0
Compilation Done!
Kriging values:
values
range 1732.05
$C_o$ 71428.57
drift equations [3, 3]
<gempy.core.interpolator.InterpolatorModel object at 0x7fcc3704b1f0>
gp.map_stack_to_surfaces(geo_data, {"Fault_Series": 'fault',
"Strat_Series": ('rock2', 'rock1')})
geo_data.set_is_fault(['Fault_Series'])
Out:
Fault colors changed. If you do not like this behavior, set change_color to False.
geo_data.get_active_grids()
Out:
array(['regular', 'topography', 'sections'], dtype='<U10')
Out:
/WorkSSD/PythonProjects/gempy/gempy/core/solution.py:173: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
self.geological_map = np.array(
/WorkSSD/PythonProjects/gempy/gempy/core/solution.py:178: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
self.sections = np.array(
Lithology ids
[4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4.
4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3.
2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4.
4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4.
3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2.
4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4.
4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3.
2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4.
4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4.
3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2.
4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4.
4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3.
2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4.
4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4.
3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2.
4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4.
4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 3. 3. 2. 2. 4. 4. 4. 4. 4. 4. 2. 2.
2. 2. 4. 4. 4. 4. 4. 4. 2. 2. 2. 2. 4. 4. 4. 4. 4. 4. 2. 2. 2. 2. 4. 4.
4. 4. 4. 4. 2. 2. 2. 2. 4. 4. 4. 4. 4. 4. 2. 2. 2. 2. 4. 4. 4. 4. 4. 4.
2. 2. 2. 2. 4. 4. 4. 4. 4. 4. 2. 2. 2. 2. 4. 4. 4. 4. 4. 4. 2. 2. 2. 2.
4. 4. 4. 4. 4. 4. 2. 2. 2. 2. 4. 4. 4. 4. 4. 4. 2. 2. 2. 2. 4. 4. 4. 4.
2. 2. 2. 2. 2. 2. 4. 4. 4. 4. 2. 2. 2. 2. 2. 2. 4. 4. 4. 4. 2. 2. 2. 2.
2. 2. 4. 4. 4. 4. 2. 2. 2. 2. 2. 2. 4. 4. 4. 4. 2. 2. 2. 2. 2. 2. 4. 4.
4. 4. 2. 2. 2. 2. 2. 2. 4. 4. 4. 4. 2. 2. 2. 2. 2. 2. 4. 4. 4. 4. 2. 2.
2. 2. 2. 2. 4. 4. 4. 4. 2. 2. 2. 2. 2. 2. 4. 4. 4. 4. 2. 2. 2. 2. 2. 2.
4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3.
2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2.
2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4.
3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2.
2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2.
4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3.
2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2.
2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4.
3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2.
2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2.
4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3.
2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2.
2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4.
3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2.
2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2.
4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2. 4. 4. 3. 3.
2. 2. 2. 2. 2. 2. 4. 4. 3. 3. 2. 2. 2. 2. 2. 2.]
new plotting api
gp.plot_2d(geo_data, section_names=['section1'])
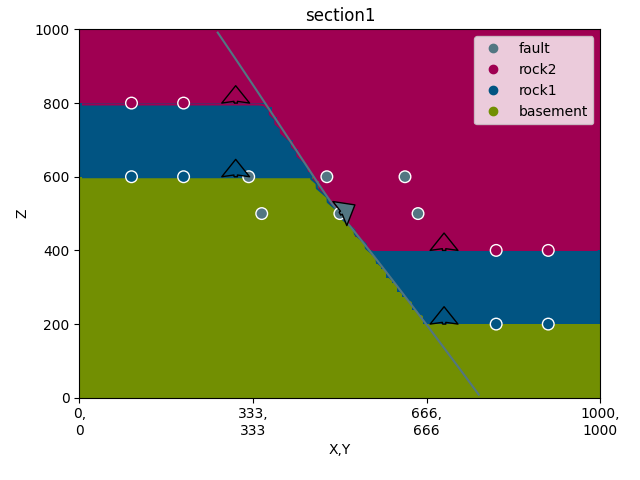
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcc748f31f0>
or
gp.plot.plot_2d(geo_data, section_names=['section1'])
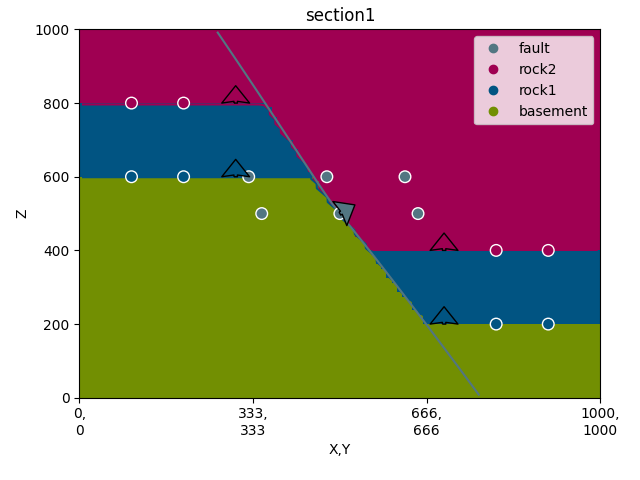
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcc3a05f580>
Plot 2d: Object oriented:¶
One plot¶
p = gp.plot_2d(geo_data, section_names=[], direction=None, show=False)
p.fig.show()

p = gp.plot_2d(geo_data, section_names=[], direction=None, show=False)
# -----new code------
sec_name = 'section1'
s1 = p.add_section(sec_name)
p.plot_data(s1, sec_name, projection_distance=200)
p.fig.show()
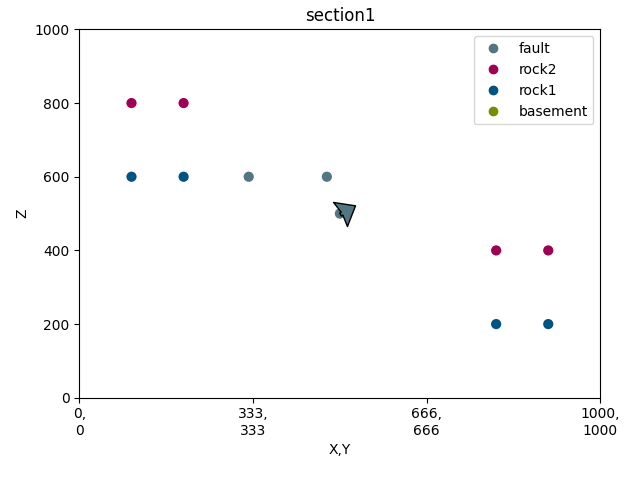
p = gp.plot_2d(geo_data, section_names=[], direction=None, show=False)
sec_name = 'section1'
s1 = p.add_section(sec_name)
# -----new code------
p.plot_data(s1, sec_name, projection_distance=200)
p.plot_contacts(s1, sec_name)
p.fig.show()
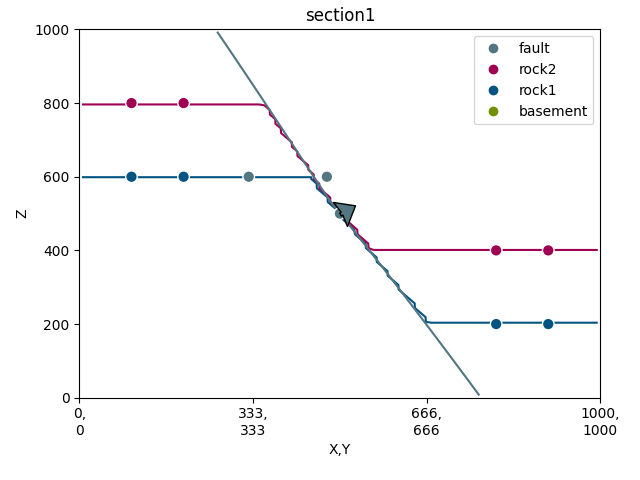
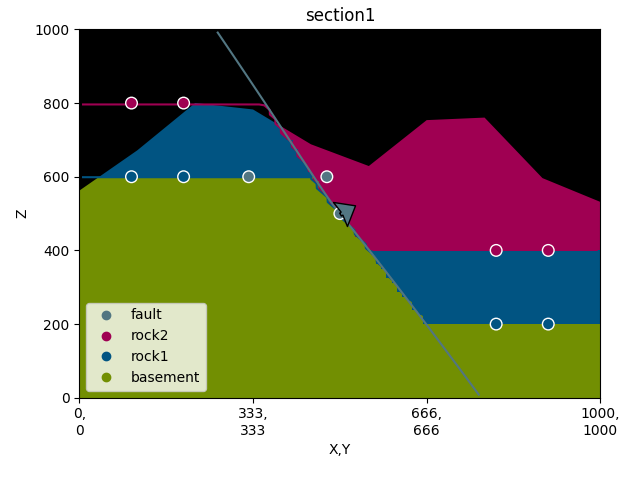
p = gp.plot_2d(geo_data, section_names=[], direction=None, show=False)
sec_name = 'section1'
s1 = p.add_section(sec_name)
p.plot_data(s1, sec_name, projection_distance=200)
p.plot_contacts(s1, sec_name)
# -----new code------
p.plot_lith(s1, sec_name)
p.plot_topography(s1, sec_name)
p.fig.show()
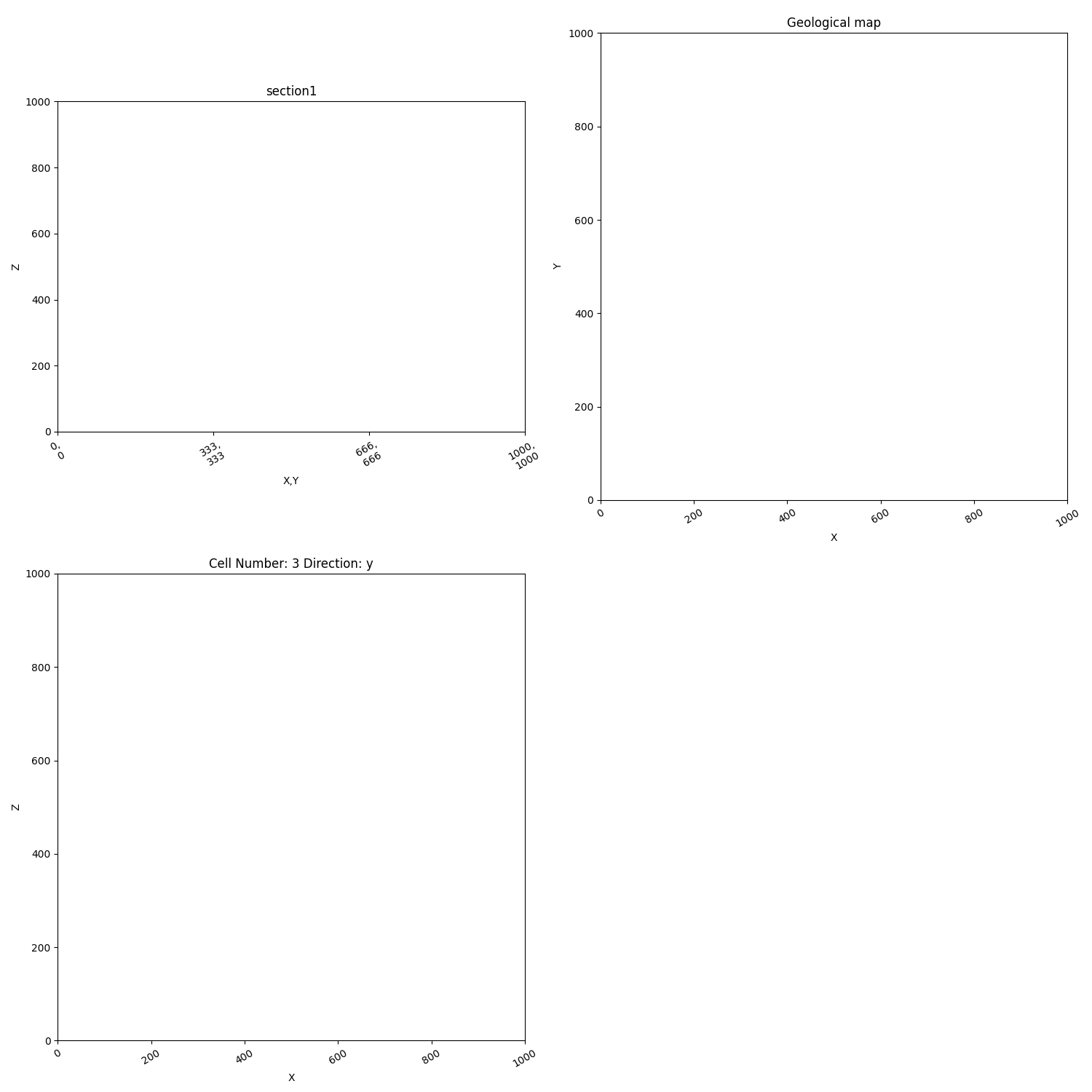
Several plots¶
sec_name = 'section1'
sec_name_2 = 'section3'
p2 = gp.plot_2d(geo_data, n_axis=3, figsize=(15, 15), # General fig options
section_names=[sec_name, 'topography'], cell_number=[3], # Defining the sections
show_data=False, show_lith=False, show_scalar=False, show_boundaries=False)
# Create the section. This loacte the axes and give the right
# aspect ratio and labels
p2 = gp.plot_2d(geo_data, n_axis=3, figsize=(15, 15), # General fig options
section_names=[sec_name, 'topography'], cell_number=[3], # Defining the sections
show_data=False, show_lith=False, show_scalar=False, show_boundaries=False,
show=False)
# -----new code------
s1 = p2.add_section(sec_name_2, ax_pos=224)
p2.fig.show()
Axes 0
p2 = gp.plot_2d(geo_data, n_axis=3, figsize=(15, 15), # General fig options
section_names=[sec_name, 'topography'], cell_number=[3], # Defining the sections
show_data=False, show_lith=False, show_scalar=False, show_boundaries=False,
show=False)
s1 = p2.add_section(sec_name_2, ax_pos=224)
# -----new code------
p2.plot_contacts(s1, sec_name_2)
p2.plot_lith(s1, sec_name_2)
p2.plot_data(s1, sec_name_2, projection_distance=200)
p2.plot_topography(s1, sec_name_2)
p2.fig.show()
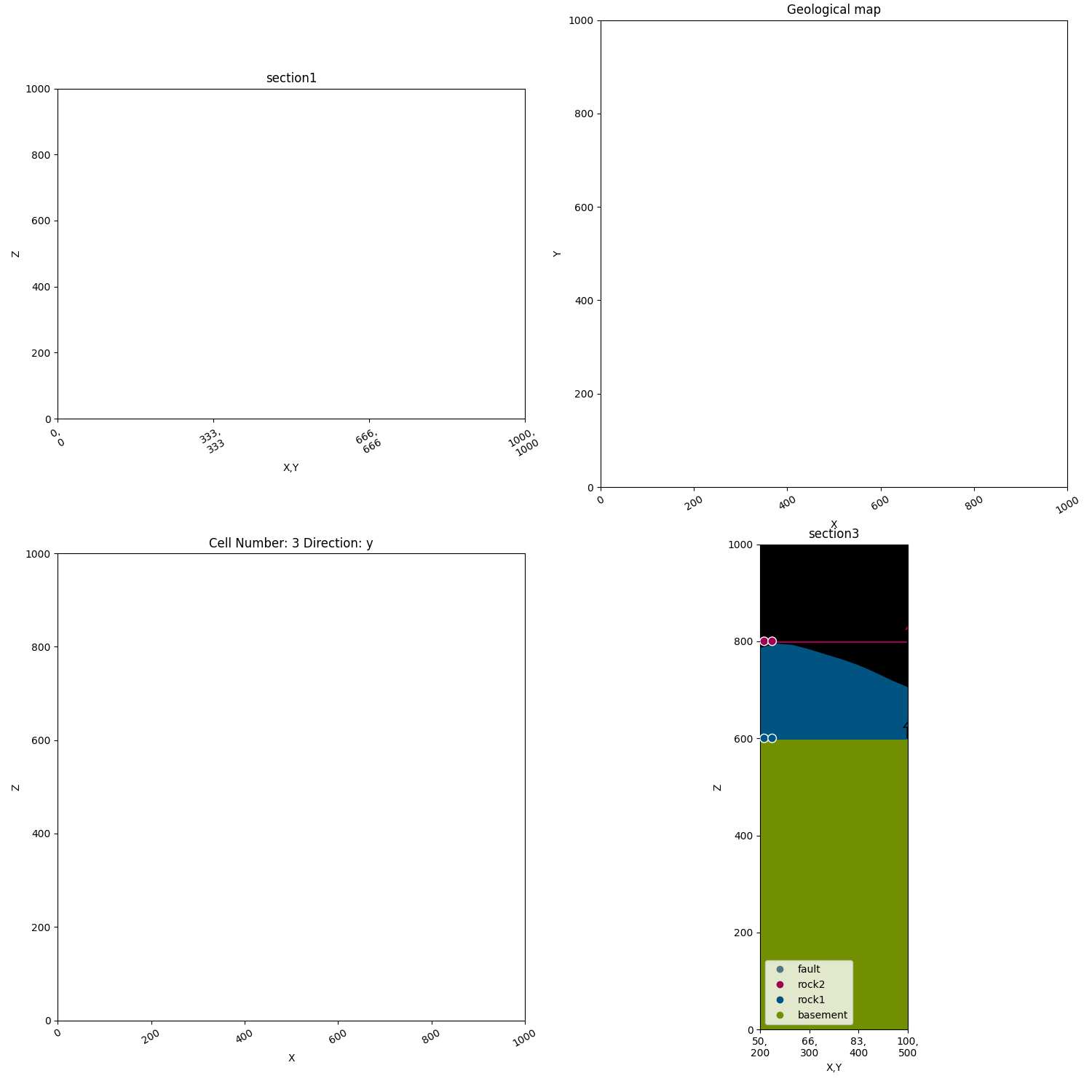
Axes 1
# sphinx_gallery_thumbnail_number = 12
p2 = gp.plot_2d(geo_data, n_axis=3, figsize=(15, 15), # General fig options
section_names=[sec_name, 'topography'], cell_number=[3], # Defining the sections
show_data=False, show_lith=False, show_scalar=False, show_boundaries=False,
show=False)
s1 = p2.add_section(sec_name_2, ax_pos=224)
p2.plot_contacts(s1, sec_name_2)
p2.plot_lith(s1, sec_name_2)
p2.plot_data(s1, sec_name_2, projection_distance=200)
p2.plot_topography(s1, sec_name_2)
# -----new code------
p2.plot_contacts(p2.axes[0], cell_number=3)
p2.plot_scalar_field(p2.axes[0], cell_number=3, series_n=1)
p2.fig.show()
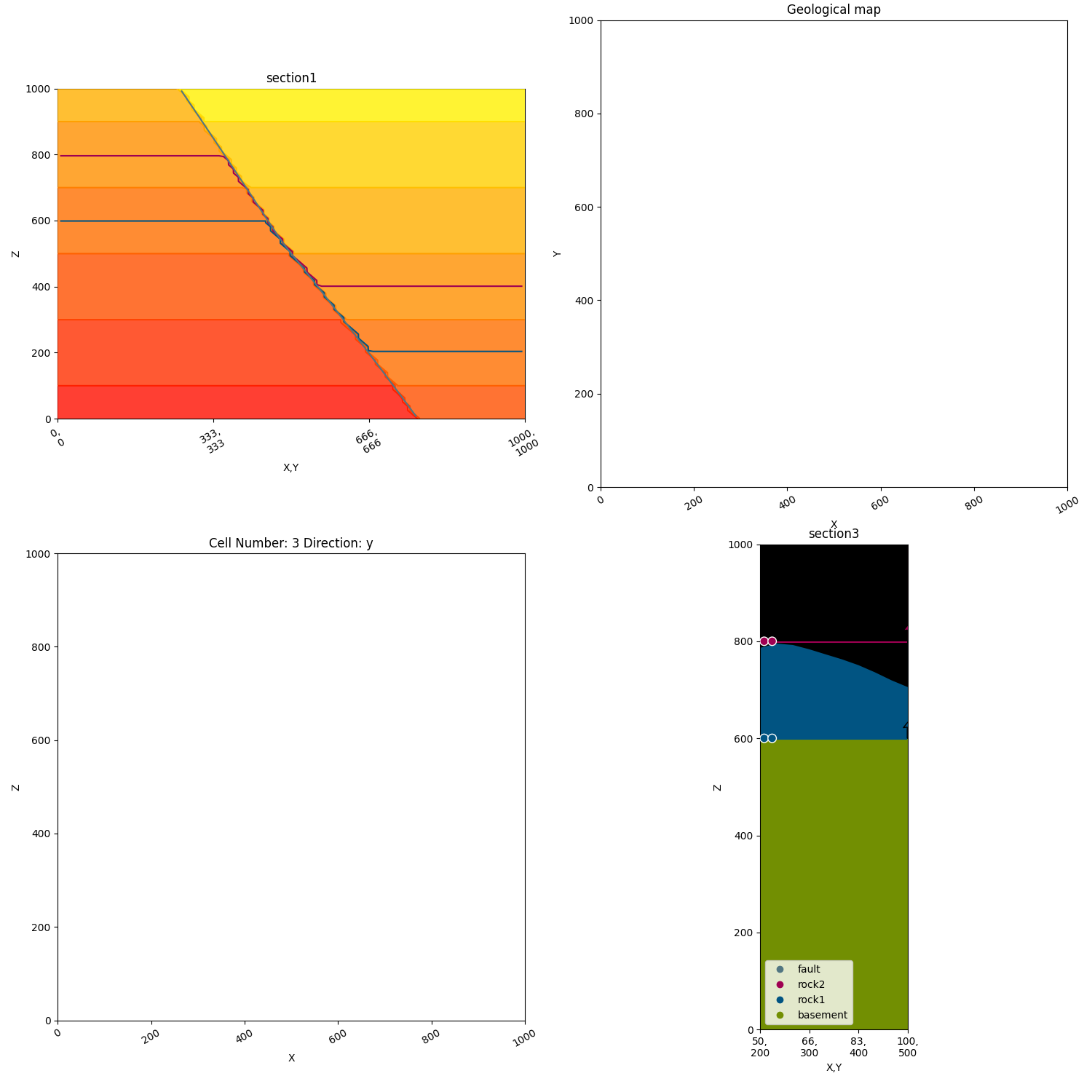
Axes2
p2 = gp.plot_2d(geo_data, n_axis=3, figsize=(15, 15), # General fig options
section_names=[sec_name, 'topography'], cell_number=[3], # Defining the sections
show_data=False, show_lith=False, show_scalar=False, show_boundaries=False,
show=False)
s1 = p2.add_section(sec_name_2, ax_pos=224)
p2.plot_contacts(s1, sec_name_2)
p2.plot_lith(s1, sec_name_2)
p2.plot_data(s1, sec_name_2, projection_distance=200)
p2.plot_topography(s1, sec_name_2)
p2.plot_contacts(p2.axes[0], cell_number=3)
p2.plot_scalar_field(p2.axes[0], cell_number=3, series_n=1)
# -----new code------
p2.plot_lith(p2.axes[1], 'topography')
p2.plot_contacts(p2.axes[1], 'topography')
p2.fig.show()
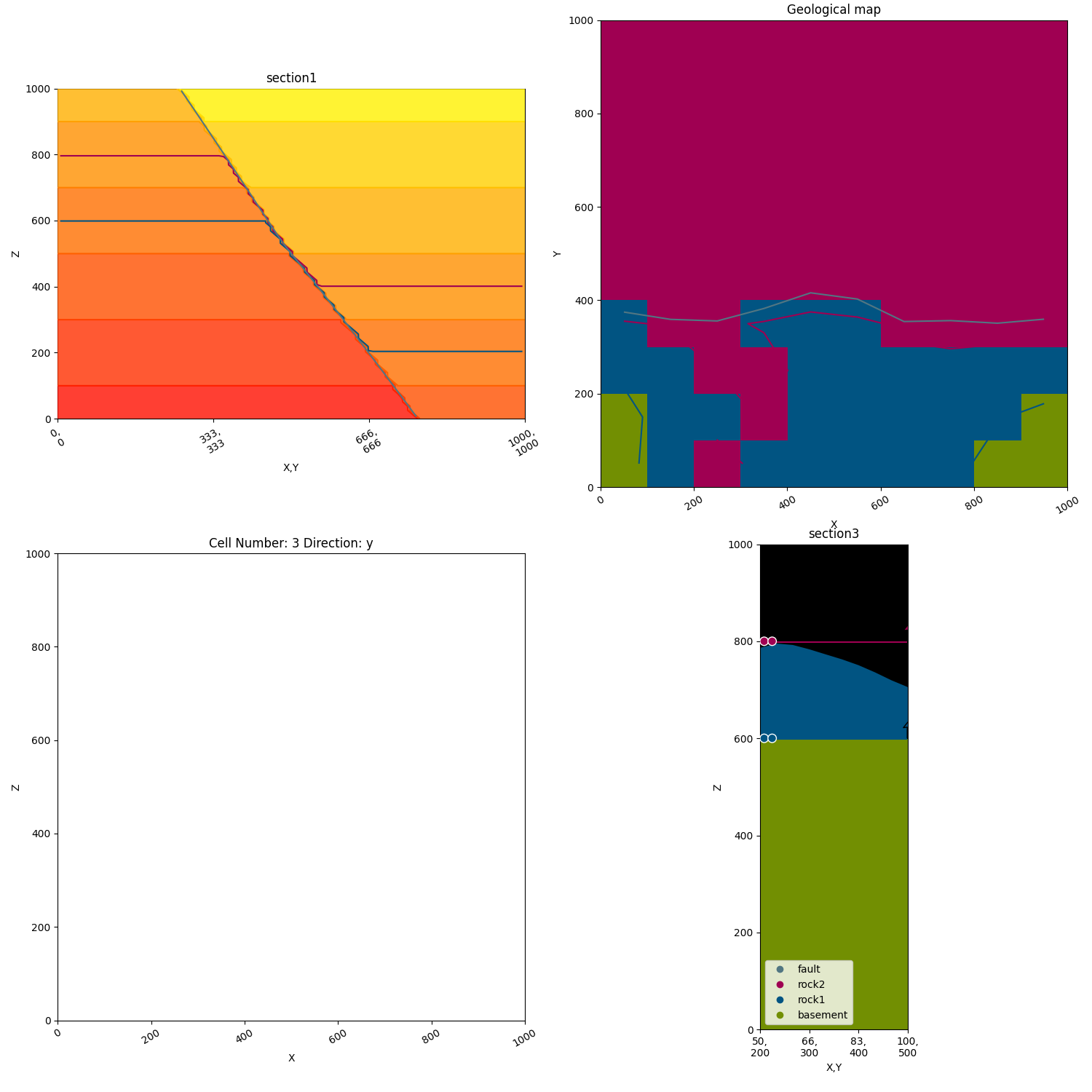
Plotting traces:¶
p2.plot_section_traces(p2.axes[1])
p2.fig.show()
gp.plot.plot_section_traces(geo_data)
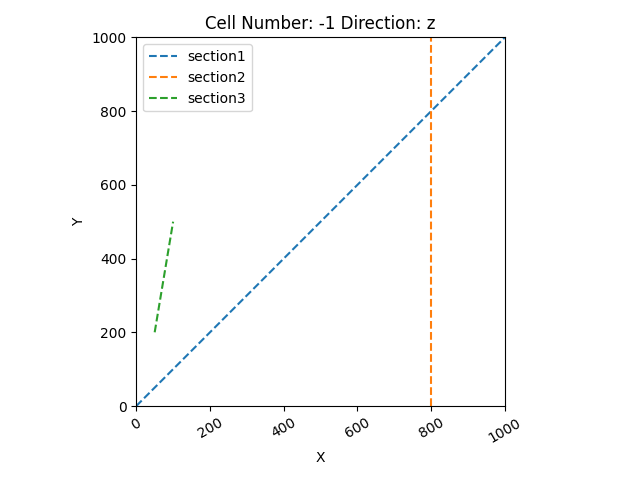
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcb84d3afd0>
Plot API¶
If nothing is passed, a Plot2D object is created and therefore you are in the same situation as above:
p3 = gp.plot_2d(geo_data)
Alternatively you can pass section_names, cell_numbers + direction or any combination of the above:
gp.plot_2d(geo_data, section_names=['topography'])
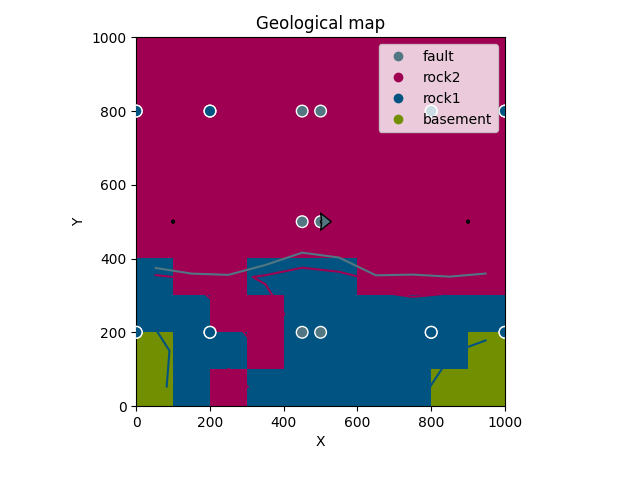
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcc46ba6910>
gp.plot_2d(geo_data, section_names=['section1'])
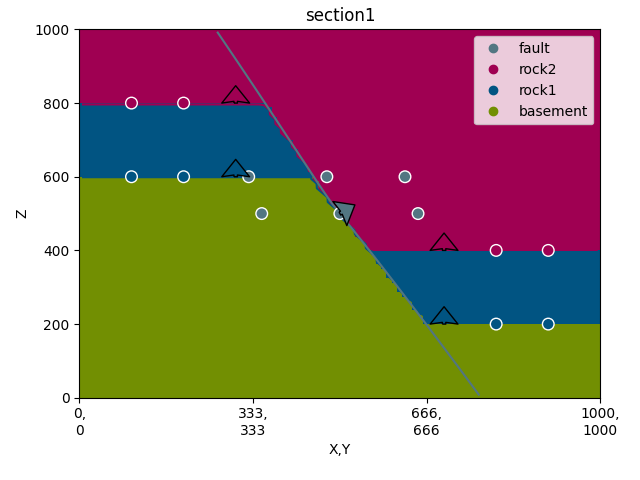
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcbe00cda90>
gp.plot_2d(geo_data, section_names=['section1', 'section2'])
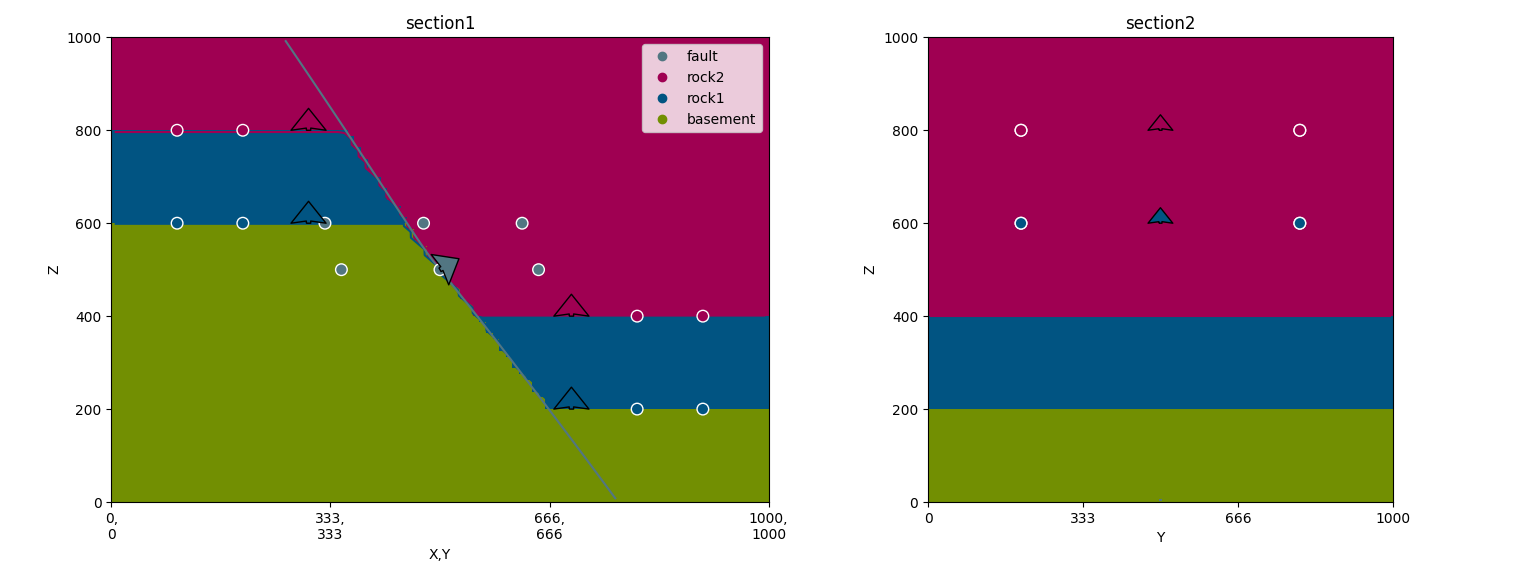
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcc46b47400>
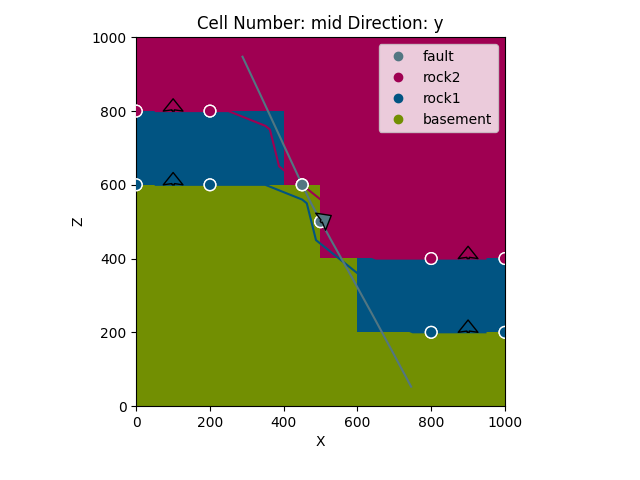
gp.plot_2d(geo_data, figsize=(15, 15), section_names=['section1', 'section2', 'topography'],
cell_number='mid')
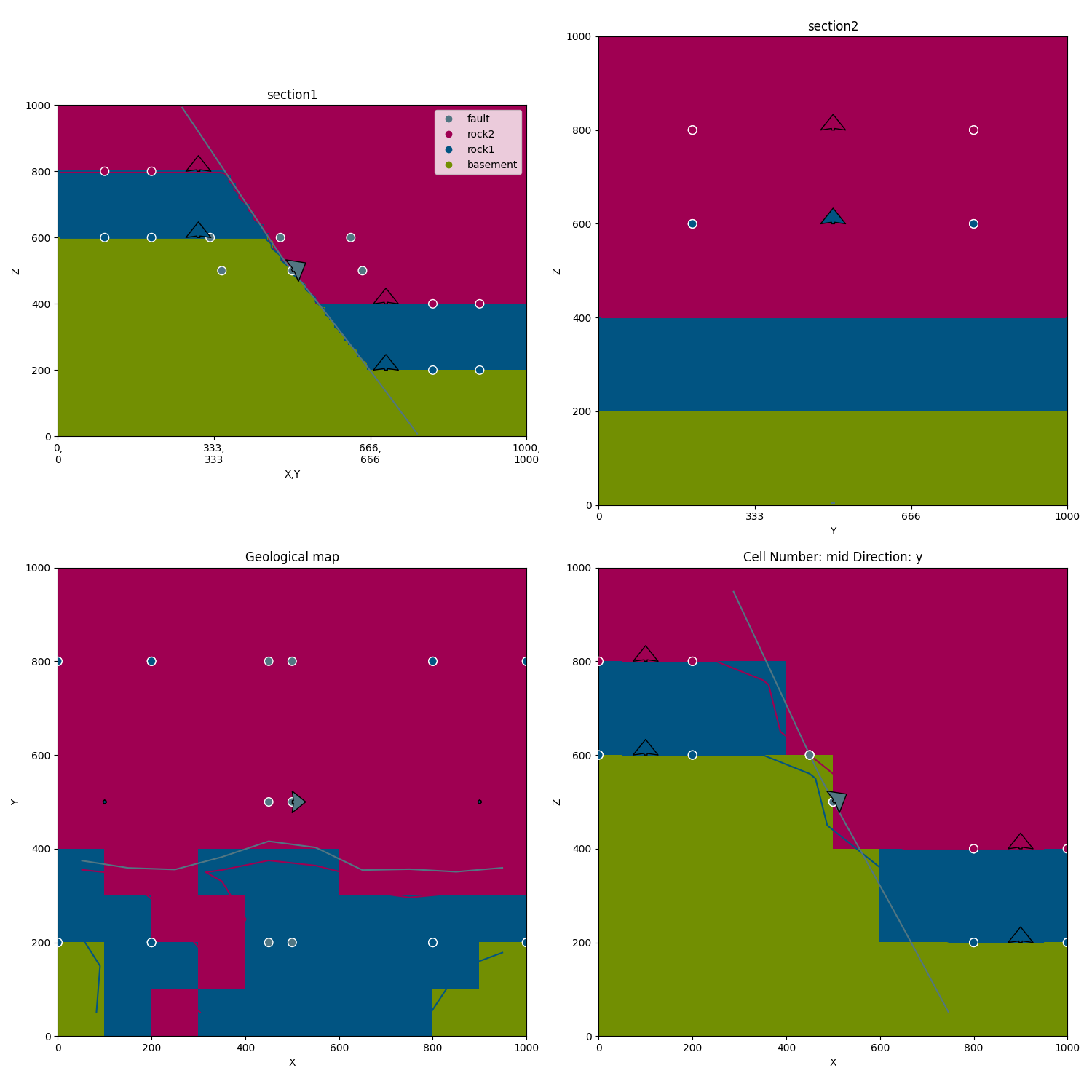
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcc74762df0>
Total running time of the script: ( 0 minutes 9.016 seconds)
